Methods of genetic analysis
The following methods can be used in genetic analysis:
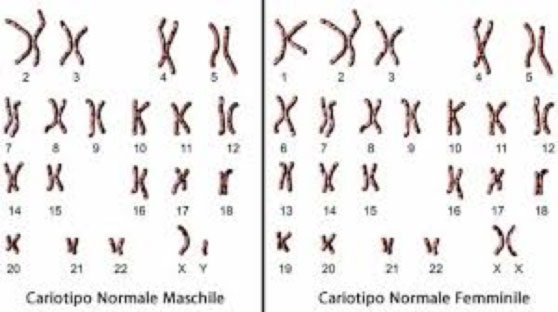
Standard chromosome examination: It is a real photograph of DNA to verify that the number of structures containing it (chromosomes) is correct. This number is characteristic of each species: the human species has 46 chromosomes organized in 23 pairs. The first 44 chromosomes form 22 pairs called “homologues” because each chromosome is received by each parent. The 23rd pair is made up of sex chromosomes, which are inherited differently from their counterparts. A female individual inherits two X chromosomes, each from a parent. A male individual, on the other hand, inherits the paternal Y chromosome and the X chromosome from the mother. There are some genetic diseases caused by anomalies in the number and / or structure of chromosomes. When the doctor suspects these conditions, the standard chromosome examination can be performed to confirm or exclude them. It is important to remember that this technique can only identify anomalies of rather large numbers, such as the lack or excess of whole chromosomes or their relevant portions. (Figure 2)
- Sanger: This technique takes its name from the geneticist who discovered it in 1977. The technique is based on the structural properties of DNA. It is a long chain made up of millions of links: each ring is called a “nucleotide”. Sanger sequencing is the reading (manual or automatic) of DNA or the set of nucleotides read in the right order. There are four types of nucleotides in the DNA, containing one of the bases: A, G, C, T. As detailed on the website https://www.emanuelemicaglio.it/category/articolo/ A and G are purines, C and T are pyrimidines. If we want to determine the sequence of a 100 nucleotide DNA fragment, it means that we want to understand what the 100 nucleotides are in the exact order. At the end of the sequencing you get a set of letters, e.g. AAGTGTGTCTGTCAGTTCTAGAACC …. In other words, the result of a Sanger sequencing allows us to determine whether and which mutation a patient is carrying. More information can be obtained from the link https://www.emanuelemicaglio.it/2019/02/18/la-genetica-medica-storia-di-un-futuro-remoto/.
- Array CGH: This is a technological improvement of the standard chromosomal examination that enables the detection of anomalies in the number of chromosomes. The CGH technique is based on the fragmentation of the patient’s DNA, which is compared with that of a certainly healthy subject of the same sex. Some heart diseases can be caused by the lack of specific fragments in chromosomes 21, 22 or on the X chromosome. Early recognition of these anomalies enables both the treatment and possible prevention of complications of heart disease.
- Next Generation Sequencing (NGS): This is a recent simplification of the Sanger technology and based on multiple and computerized DNA reading. It allows the study of a “panel” of genes at one time, comprising dozens of DNA fragments, each of which can be read hundreds and hundreds of times. In Cardiogenetics, it is common that the same disease can be caused by many different mutations, in different genes. Therefore, NGS technology has revolutionized our specialty, allowing in many cases to obtain confirmation or exclusion of a disease that was clinically suspected.
- MLPA: This is an in-depth examination that is generally used when sequencing (Sanger or NGS) is negative, but clinically there is a well-founded suspicion of a genetic condition. The technique is similar to that of CGH and allows to check whether the gene (= DNA fragment) being tested is present in two copies. Otherwise, two things can occur: no copy (= gene deletion) or three copies (= gene duplication). In both of these cases, the consequence for the patient may be heart disease. It should be noted that a mechanism such as the loss or gain of a gene can cause complex and generally syndromic heart disease (i.e. with involvement of other organs besides the heart).
Whole Genome Sequencing (WGS): This enables the examination of the entire genetic heritage of a patient and, at the moment (September 2019), has limited use for research. The examination is based on the same technique as the NGS, but also includes regions of DNA not yet studied (Figure 3). Our group analyzes the samples with the WGS method only in patients with Brugada syndrome. Therefore, the test studies not only the genes currently known to be associated with the Brugada syndrome, but also other genes whose function is still yet unknown. Therefore, patients who adhere to the research protocol are carefully informed about the possible implications of the results for their health. Each patient can decide to know only the cause of his condition or to also know any unexpected results, which could have enormous importance for their health.

If you want to know more >Cardiogenetics
